ABSTRACT
There is a pressing need to identify therapeutic targets in tumors with low mutation rates such as the malignant pediatric brain tumor medulloblastoma. To address this challenge, we quantitatively profiled global proteomes and phospho-proteomes of 45 medulloblastoma samples. Integrated analyses revealed that tumors with similar RNA expression vary extensively at the post-transcriptional and post-translational levels. We identified distinct pathways associated with two subsets of SHH tumors, and found post-translational modifications of MYC that are associated with poor outcomes in group 3 tumors. We found kinases associated with subtypes and showed that inhibiting PRKDC sensitizes MYC-driven cells to radiation. Our study shows that proteomics enables a more comprehensive, functional readout, providing a foundation for future therapeutic strategies.
Data & Availability
- The accession number for the proteomic, phospho-proteomic, and acetylome data reported in this paper is MSV000082644
- Group 3 cohort expansion tool reported in the paper can be located at https://github.com/ckmah/archer2018_g3_cohort_expansion/.
- The genomic data used in this paper is a subset of the Northcott et al. 2017 dataset, which can be found at https://www.nature.com/articles/nature22973.
- To browse and explore the data from this study go to http://fraenkel.mit.edu/archer2018
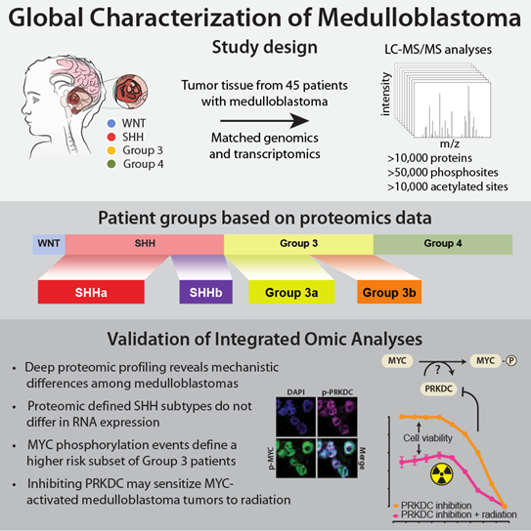
Highlights
- Deep proteomic profiling reveals mechanistic differences among medulloblastomas
- Proteomic-defined SHH subtypes do not differ in RNA expression
- MYC phosphorylation events define a higher risk subset of group 3 patients
- Inhibiting PRKDC may sensitize MYC-activated medulloblastoma tumors to radiation
Significance
Genomic and epigenomic analyses have revolutionized cancer diagnostics. Nevertheless, it has been difficult to identify therapeutic targets for tumors that lack recurrent genomic lesions. Here we used global, mass spectrometry-based measurements of protein levels and post-translational modifications to identify functional pathways associated with subtypes of medulloblastoma. Strong proteomic signals revealed altered pathways that were not detected transcriptionally. One molecular subgroup of tumors showed marked discordance of RNA and protein levels, suggesting global changes in translation and/or proteostasis. We demonstrate the utility of an integrative approach for discovery of candidate biomarkers or drug targets and provide a multi-omic dataset that will serve as a resource for the community. This study has the potential to impact clinical trial design.